Abstract
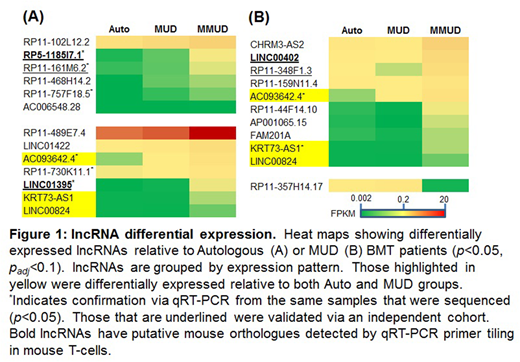
The mechanisms governing human allogeneic T cell responses remain incompletely understood which limits the wider clinical application of allo-BMT. Long non-coding RNAs (lncRNA) are an emerging class of non-coding RNAs that control gene expression with tissue and context specificity. However, their role in alloimmunity is unknown. To determine whether lncRNAs regulate T cell alloimmunity we performed RNA-seq of donor T cells following clinical BMT to identify and characterize differentially-expressed lncRNAs. CD3+ T cells 30 days post myeloablative BMT were isolated in a discovery cohort from 3 different groups, each with increasing amounts of allo-antigen stimulation: 1) autologous donors (Auto) to control for lymphopenia induced proliferation; 2) matched unrelated donors (MUD), which receive minor histocompatibility allo-stimulation and are exposed to immune suppression; and 3) mis-matched unrelated donors (MMUD), which receive both minor and major histocompatibility allo-stimulation and also receive immune suppression. Subjects with GVHD, infection, who received corticosteroids, or who received T cell depleting therapy were excluded. Hierarchical clustering showed distinct grouping of MMUD samples, but an intermingling of the Auto and MUD samples suggesting that increasing allogeneic stimulation drove differential transcript expression. Gene ontology analysis of protein coding genes showed enrichment for pathways operant in activated T cells, thereby providing validity to our approach. The differentially expressed lncRNAs mainly showed increasing expression with increasing allo-stimulation (i.e. higher expression in the MMUD group) (Figure 1). We validated the sequencing data with qRT-PCR for 6 of 8 lncRNAs with sequencing padj < 0.05 relative to the Auto group (see those marked with an * in Figure 1).
To further validate the sequencing data, we confirmed the differential expression of 5 lncRNAs in an independent patient cohort (n=8-11 per group) by qRT-PCR (see those underlined in Figure 1). Linc00402 was chosen as the lead candidate (increased 3 fold in the MMUD group, p < 0.01). Its expression along with the 4 other lncRNAs were further confirmed to be regulated by allo-stimulation following in vitro human mixed lymphocyte reactions (MLRs) (Linc00402's expression changed 4.7 fold, p < 0.001). The database FANTOM-CAT showed strong enrichment of Linc00402 (88 fold) in human T cells relative to other cell/tissue types, thereby highlighting its tissue specific expression. This was validated in CD4 and CD8 human T cells which demonstrated similar expression of Linc00402.
We next explored for murine orthologues of Linc00402 and the other 4 differentially expressed lncRNAs using Transmap. Expression of 3 of the 4 putative orthologues that were identified by Transmap was detected by qRT-PCR primer tiling (see those in bold in Figure 1). Their differential expression upon allo-stimulation, including the putative Linc00402 orthologue, GM34199, was confirmed in vitro with an MLR (22 fold change in expression of GM34199, p < 0.01) and in vivo using both major and minor histocompatibility disparate murine models of GVHD (B6gBalb/c and B6gC3H.SW, respectively) at 7 and 14 days post BMT in which GM34199's expression changed 2.5 to 13 fold (p < 0.01).
The functional validation of human Linc00402 and murine GM34199 was tested by anti-sense oligonucleotide-mediated knockdown, which inhibited both human (55% of control, p < 0.01) and murine (32% of control, p < 0.01) allogeneic T cell proliferation. Further mechanistic studies revealed that Linc00402 is predominantly expressed in the cytoplasm following subcellular fractionation.
These results indicate that Linc00402 in humans and its putative murine orthologue are differentially expressed by allogeneic T cells and regulate their function. In light of its tissue and allogeneic context specific expression, Linc00402 could be an attractive target for improving outcomes after allogeneic BMT.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal